Automatic Control Knowledge Repository
You currently have javascript disabled. Some features will be unavailable. Please consider enabling javascript.Details for: "lotka volterra"
Name: lotka volterra
(Key: DRZGT)
Path: ackrep_data/system_models/lotka_volterra View on GitHub
Type: system_model
Short Description: predator-prey model, used to describe the population dynamics of biological systems
Created: 05.08.2022
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
Related Problems:
SINDy for Lotka Volterra Model
Extensive Material:
Download pdf
Result: Success.
Last Build: Checkout CI Build
Runtime: 3.2 (estimated: 10s)
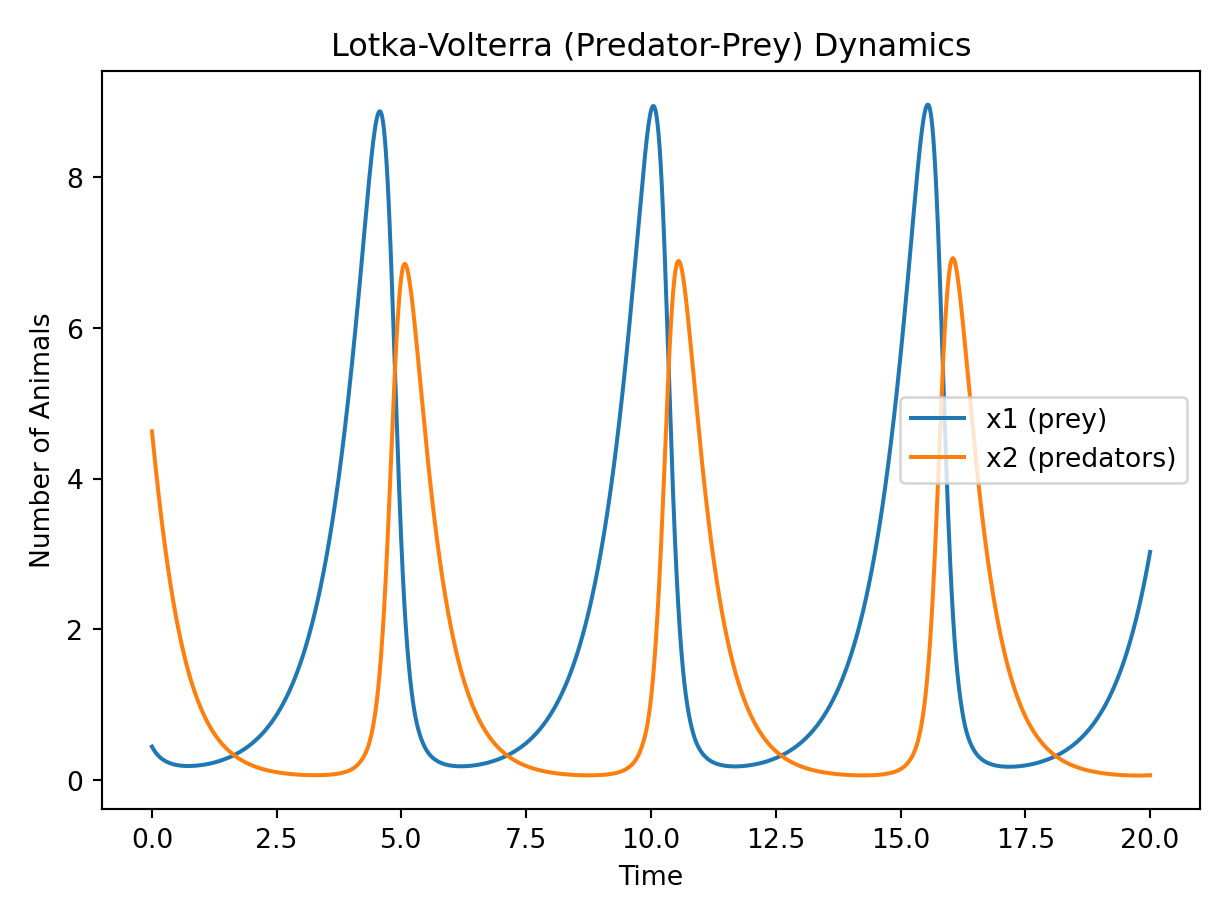
Plot:
The image of the latest CI job is not available. This is a fallback image.
Path: ackrep_data/system_models/lotka_volterra View on GitHub
Type: system_model
Short Description: predator-prey model, used to describe the population dynamics of biological systems
Created: 05.08.2022
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
from cProfile import label
import numpy as np
import system_model
from scipy.integrate import solve_ivp
from ackrep_core import ResultContainer
from ackrep_core.system_model_management import save_plot_in_dir
import matplotlib.pyplot as plt
import os
# link to documentation with examples: https://ackrep-doc.readthedocs.io/en/latest/devdoc/contributing_data.html
def simulate():
"""
simulate the system model with scipy.integrate.solve_ivp
:return: result of solve_ivp, might contains input function
"""
model = system_model.Model()
rhs_xx_pp_symb = model.get_rhs_symbolic()
print("Computational Equations:\n")
for i, eq in enumerate(rhs_xx_pp_symb):
print(f"dot_x{i+1} =", eq)
rhs = model.get_rhs_func()
# initial state values
xx0 = [0.44249296, 4.6280594]
t_end = 20
tt = np.linspace(0, t_end, 10000)
simulation_data = solve_ivp(rhs, (0, t_end), xx0, t_eval=tt)
print(simulation_data.y[:, -1])
save_plot(simulation_data)
return simulation_data
def save_plot(simulation_data):
"""
plot your data and save the plot
access to data via: simulation_data.t array of time values
simulation_data.y array of data components
simulation_data.uu array of input values
:param simulation_data: simulation_data of system_model
:return: None
"""
# plot of your data
plt.plot(simulation_data.t, simulation_data.y[0], label="$x_1$ (prey)")
plt.plot(simulation_data.t, simulation_data.y[1], label="$x_2$ (predators)")
plt.legend()
plt.grid()
plt.xlabel("Time [s]")
plt.ylabel("Number of Animals")
plt.title("Lotka-Volterra (Predator-Prey) Dynamics")
plt.tight_layout()
save_plot_in_dir()
def evaluate_simulation(simulation_data):
"""
assert that the simulation results are as expected
:param simulation_data: simulation_data of system_model
:return:
"""
expected_final_state = [3.02762979, 0.06256621]
rc = ResultContainer(score=1.0)
simulated_final_state = simulation_data.y[:, -1]
rc.final_state_errors = [
simulated_final_state[i] - expected_final_state[i] for i in np.arange(0, len(simulated_final_state))
]
rc.success = np.allclose(expected_final_state, simulated_final_state, rtol=0, atol=1e-2)
return rc
import sympy as sp
import symbtools as st
import importlib
import sys, os
# from ipydex import IPS, activate_ips_on_exception
from ackrep_core.system_model_management import GenericModel, import_parameters
# Import parameter_file
params = import_parameters()
# link to documentation with examples: https://ackrep-doc.readthedocs.io/en/latest/devdoc/contributing_data.html
class Model(GenericModel):
def initialize(self):
"""
this function is called by the constructor of GenericModel
:return: None
"""
# ---------start of edit section--------------------------------------
# Define number of inputs -- MODEL DEPENDENT
self.u_dim = 0
# Set "sys_dim" to constant value, if system dimension is constant
self.sys_dim = 2
# ---------end of edit section----------------------------------------
# check existence of params file
self.has_params = True
self.params = params
def get_rhs_symbolic(self):
"""
define symbolic rhs function
:return: matrix of symbolic rhs-functions
"""
if self.dxx_dt_symb is not None:
return self.dxx_dt_symb
# ---------start of edit section--------------------------------------
x1, x2 = self.xx_symb # state components
a, b, c, d = self.pp_symb # parameters
# define symbolic rhs functions
dx1_dt = a * x1 - b * x1 * x2
dx2_dt = c * x1 * x2 - d * x2
# rhs functions matrix
self.dxx_dt_symb = sp.Matrix([dx1_dt, dx2_dt])
# ---------end of edit section----------------------------------------
return self.dxx_dt_symb
import sys
import os
import numpy as np
import sympy as sp
import tabulate as tab
# link to documentation with examples: https://ackrep-doc.readthedocs.io/en/latest/devdoc/contributing_data.html
# set model name
model_name = "Lotka_Volterra"
# ---------- create symbolic parameters
pp_symb = [a, b, c, d] = sp.symbols("a, b, c, d", real=True)
# set numerical values of auxiliary parameters
# tailing "_nv" stands for "numerical value"
a_nv = 1.3
b_nv = 0.9
c_nv = 0.8
d_nv = 1.8
# list of symbolic parameter functions
# tailing "_sf" stands for "symbolic parameter function"
pp_sf = [a_nv, b_nv, c_nv, d_nv]
# OPTIONAL
# range of parameters
a_range = (0, np.inf)
b_range = (0, np.inf)
c_range = (0, np.inf)
d_range = (0, np.inf)
# OPTIONAL
# list of ranges
pp_range_list = [a_range, b_range, c_range, d_range]
# ---------- list for substitution
# -- entries are tuples like: (independent symbolic parameter, numerical value)
pp_subs_list = []
# OPTONAL: Dictionary which defines how certain variables shall be written
# in the table - key: Symbolic Variable, Value: LaTeX Representation/Code
# useful for example for complex variables: {Z: r"\underline{Z}"}
latex_names = {a: r"\alpha", b: r"\beta", c: r"\gamma", d: r"\delta"}
# ---------- Define LaTeX table
# ---------- CREATE BEGIN OF LATEX TABULAR
# Define tabular Header
# DON'T CHANGE FOLLOWING ENTRIES: "Symbol", "Value"
tabular_header = ["Parameter Name", "Symbol", "Value"]
# Define column text alignments
col_alignment = ["left", "center", "left"]
# Define Entries of all columns before the Symbol-Column
# --- Entries need to be latex code
col_1 = [
"reproduction rate of prey alone",
"mortality rate of prey per predator",
"mortality rate of predators",
"reproduction rate of predators per prey",
]
# contains all lists of the columns before the "Symbol" Column
# --- Empty list, if there are no columns before the "Symbol" Column
start_columns_list = [col_1]
# contains all lists of columns after the FIX ENTRIES
# --- Empty list, if there are no columns after the "Value" column
end_columns_list = []
Related Problems:
SINDy for Lotka Volterra Model
Extensive Material:
Download pdf
Result: Success.
Last Build: Checkout CI Build
Runtime: 3.2 (estimated: 10s)
Plot:

The image of the latest CI job is not available. This is a fallback image.