Automatic Control Knowledge Repository
You currently have javascript disabled. Some features will be unavailable. Please consider enabling javascript.Details for: "modular multilevel converter (MMC)"
Name: modular multilevel converter (MMC)
(Key: YHS5B)
Path: ackrep_data/system_models/mmc_system View on GitHub
Type: system_model
Short Description: MMC with 3 phases, 1 Submodule per Arm and output to grid. Used to generate multilevel Voltages
Created: 2022-05-09
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
Related Problems:
Extensive Material:
Download pdf
Result: Success.
Last Build: Checkout CI Build
Runtime: 39.6 (estimated: 50s)
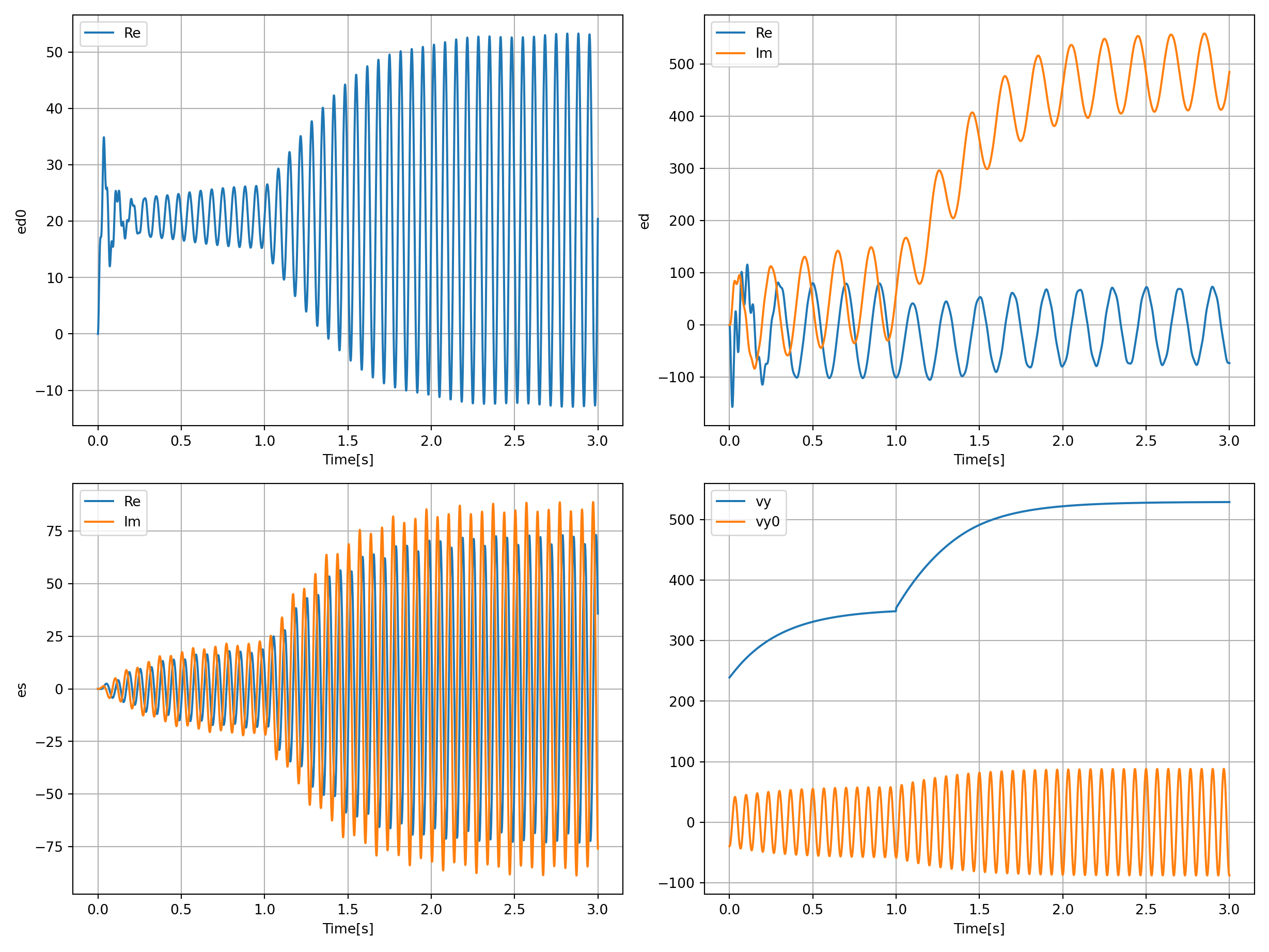
Plot:
The image of the latest CI job is not available. This is a fallback image.
Path: ackrep_data/system_models/mmc_system View on GitHub
Type: system_model
Short Description: MMC with 3 phases, 1 Submodule per Arm and output to grid. Used to generate multilevel Voltages
Created: 2022-05-09
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
import numpy as np
import system_model
from scipy.integrate import solve_ivp
from ackrep_core import ResultContainer
from ackrep_core.system_model_management import save_plot_in_dir
import matplotlib.pyplot as plt
import os
import time
from ipydex import IPS, activate_ips_on_exception
activate_ips_on_exception()
def simulate():
model = system_model.Model()
rhs_xx_pp_symb = model.get_rhs_symbolic()
print("Computational Equations:\n")
for i, eq in enumerate(rhs_xx_pp_symb):
print(f"dot_x{i+1} =", eq)
rhs = model.get_rhs_func()
# --------------------------------------------------------------------
xx0 = [0, 0, 0 + 0j, 0 + 0j, 0 + 0j, 0, 0 + 0j, 0]
t_end = 3
tt = np.linspace(0, t_end, 3000)
# use model class rhs
sol = solve_ivp(rhs, (0, t_end), xx0, t_eval=tt)
i = 0
n_rows = len(sol.t)
n_cols = 4
uu = np.zeros((n_rows, n_cols))
while i < len(sol.t):
uu[i, :] = model.uu_func(sol.t[i], sol.y[:, i])
i = i + 1
sol.uu = uu
# --------------------------------------------------------------------
save_plot(sol)
return sol
def save_plot(sol):
# --------------------------------------------------------------------
# create figure + 2x2 axes array
fig1, axs = plt.subplots(nrows=2, ncols=2, figsize=(12.8, 9.6))
# print in axes top left
axs[0, 0].plot(sol.t, np.real(sol.y[1]), label="Re")
axs[0, 0].set_ylabel("ed0") # y-label
axs[0, 0].set_xlabel("Time [s]") # x-Label
axs[0, 0].grid()
axs[0, 0].legend()
# print in axes top right
axs[1, 0].plot(sol.t, np.real(sol.y[2]), label="Re")
axs[1, 0].plot(sol.t, np.imag(sol.y[2]), label="Im")
axs[1, 0].set_ylabel("es") # y-label
axs[1, 0].set_xlabel("Time [s]") # x-Label
axs[1, 0].grid()
axs[1, 0].legend()
# print in axes bottom left
axs[0, 1].plot(sol.t, np.real(sol.y[3]), label="Re")
axs[0, 1].plot(sol.t, np.imag(sol.y[3]), label="Im")
axs[0, 1].set_ylabel("ed") # y-label
axs[0, 1].set_xlabel("Time [s]") # x-Label
axs[0, 1].grid()
axs[0, 1].legend()
# print in axes bottom right
axs[1, 1].plot(sol.t, sol.uu[:, 0], label="vy")
axs[1, 1].plot(sol.t, sol.uu[:, 1], label="vy0")
axs[1, 1].set_ylabel("") # y-label
axs[1, 1].set_xlabel("Time [s]") # x-Label
axs[1, 1].grid()
axs[1, 1].legend()
# adjust subplot positioning and show the figure
fig1.subplots_adjust(hspace=0.5)
# fig1.show()
# --------------------------------------------------------------------
plt.tight_layout()
save_plot_in_dir()
def evaluate_simulation(simulation_data):
"""
:param simulation_data: simulation_data of system_model
:return:
"""
# --------------------------------------------------------------------
# fill in final states of simulation to check your model
# simulation_data.y[i][-1]
expected_final_state = [
(55.43841456212572 + 0j),
(20.413747733428224 + 0j),
(35.79509623855708 - 75.96575244375583j),
(-72.91879818170683 + 485.1484950343515j),
0j,
(17.557881788491308 + 0j),
(9.991045345136369 - 3.6215387749437133j),
(94.24777960769383 + 0j),
]
# --------------------------------------------------------------------
rc = ResultContainer(score=1.0)
simulated_final_state = simulation_data.y[:, -1]
rc.final_state_errors = [
simulated_final_state[i] - expected_final_state[i] for i in np.arange(0, len(simulated_final_state))
]
rc.success = np.allclose(expected_final_state, simulated_final_state, rtol=0, atol=1e-2)
return rc
import sympy as sp
import symbtools as st
import importlib
from sympy import I
import numpy as np
import sys, os
from ipydex import IPS, activate_ips_on_exception # for debugging only
from ackrep_core.system_model_management import GenericModel, import_parameters
# Import parameter_file
params = import_parameters()
class Model(GenericModel):
def initialize(self):
"""
this function is called by the constructor of GenericModel
:return: None
"""
# Define number of inputs -- MODEL DEPENDENT
self.u_dim = 4
# Set "sys_dim" to constant value, if system dimension is constant
# else set "sys_dim" to x_dim -- MODEL DEPENDENT
self.sys_dim = 8
# check existance of params file
self.has_params = True
self.params = params
# ----------- SET DEFAULT INPUT FUNCTION ---------- #
# --------------- Only for non-autonomous Systems
# --------------- MODEL DEPENDENT
def uu_default_func(self):
"""
:return: (function with 2 args - t, xx_nv) default input function
"""
vdc, vg, omega, Lz, Mz, R, L = list(self.pp_dict.values())
Ind_sum = Mz + Lz
def uu_rhs(t, xx_nv):
"""
:param t:(scalar or vector) Time
:param xx_nv: (vector or array of vectors) state vector with
numerical values at time t
:return: [vy, vy0, vx, vx0]
"""
es0, ed0, es, ed, iss, iss0, i, theta = xx_nv
# Define Input Functions
Kp = 5
# define reference trajectory for i
T_dur = 1.5
tau = t / T_dur
i_max = 10
i_ref = 10
dt_i_ref = 0
if tau < 1:
i_ref = 4 + (i_max - 4) * np.sin(0.5 * tau * np.pi) * np.sin(0.5 * tau * np.pi)
dt_i_ref = np.pi / T_dur * np.sin(0.5 * tau * np.pi) * np.cos(0.5 * tau * np.pi)
if t < 1:
i_ref = 4
dt_i_ref = 0
vy = vg + (R + 1j * omega * L) * i + 1 * (i_ref - i) + L * dt_i_ref
vy = complex(vy)
vy0 = -1 / 6 * np.absolute(vy) * np.real(np.exp(3 * 1j * (theta + np.angle(vy))))
vx = 1j * omega * Ind_sum * iss - Kp * iss # p- controller to get iss
# define reference trajectory of es0
T_dur = 0.5 # duration of es0 ref_traj
tau = t / T_dur
# es0_max = 68
# es0_ref = sp.Piecewise((0, tau < 0), (es0_max *sp.sin( tau*sp.pi/2) \
# *sp.sin( tau*sp.pi/2), tau < 1), (es0_max, True) )
es0_ref = 56
# derive iss0 ref trajectory
iss0_ref = 1 / vdc * np.real(i * np.conjugate(vy)) + Kp * (es0_ref - es0)
# p-controller for vx0
vx0 = Kp * (iss0_ref - iss0)
return [np.real(vy), vy0, vx, vx0]
return uu_rhs
# ----------- SYMBOLIC RHS FUNCTION ---------- #
# --------------- MODEL DEPENDENT
def get_rhs_symbolic(self):
"""
:return:(matrix) symbolic rhs-functions
"""
if self.dxx_dt_symb is not None:
return self.dxx_dt_symb
es0, ed0, es, ed, iss, iss0, i, theta = self.xx_symb
vdc, vg, omega, Lz, Mz, R, L = self.pp_symb
# u0 = input force
vy0, vy, vx0, vx = self.uu_symb
# Auxiliary variables
Ind_sum = Lz + Mz
vydelta = vy - Mz * (I * omega * i + 1 / L * (vy - (R + I * omega * L) * i - vg))
# create symbolic rhs functions
des0_dt = vdc * iss0 - sp.re(i * sp.conjugate(vy))
ded0_dt = -2 * vy0 * iss0 - sp.re(sp.conjugate(iss) * vydelta)
des_dt = vdc * iss - sp.exp(-3 * I * theta) * sp.conjugate(vy) * sp.conjugate(i) - 2 * i * vy0 - I * omega * es
ded_dt = (
vdc * i
- sp.exp(-3 * I * theta) * sp.conjugate(iss) * sp.conjugate(vydelta)
- 2 * iss * vy0
- 2 * iss0 * vydelta
- I * omega * ed
)
diss_dt = 1 / Ind_sum * (vx - I * omega * Ind_sum * iss)
diss0_dt = vx0 / Ind_sum
di_dt = 1 / L * (vy - (R + I * omega * L) * i - vg)
dtheta_dt = 2 * sp.pi / 360 * omega
# put rhs functions into a vector
self.dxx_dt_symb = sp.Matrix([des0_dt, ded0_dt, des_dt, ded_dt, diss_dt, diss0_dt, di_dt, dtheta_dt])
return self.dxx_dt_symb
# ----------- NUMERIC RHS FUNCTION ---------- #
# -------------- written sepeeratly cause it seems like that lambdify can't
# -------------- handle complex values in a proper way
def get_rhs_func(self):
"""
Creates an executable function of the model which uses:
- the current parameter values
- the current input function
To evaluate the effect of a changed parameter set a new rhs_func needs
to be created with this method.
:return:(function) rhs function for numerical solver like
scipy.solve_ivp
"""
# create rhs function
def rhs(t, xx_nv):
"""
:param t:(tuple or list) Time
:param xx_nv:(self.n-dim vector) numerical state vector
:return:(self.n-dim vector) first time derivative of state vector
"""
uu_nv = self.uu_func(t, xx_nv)
vy, vy0, vx, vx0 = uu_nv
es0, ed0, es, ed, iss, iss0, i, theta = xx_nv
vdc, vg, omega, Lz, Mz, R, L = list(self.pp_dict.values())
Ind_sum = Mz + Lz
# = vy-Mz(j*omega*i-dt_i)
vydelta = vy - Mz * (1j * omega * i + 1 / L * (vy - (R + 1j * omega * L) * i - vg))
dt_es0 = vdc * iss0 - np.real(i * np.conjugate(vy))
dt_ed0 = -2 * vy0 * iss0 - np.real(np.conjugate(iss) * vydelta)
dt_es = (
vdc * iss - np.exp(-3 * 1j * theta) * np.conjugate(vy) * np.conjugate(i) - 2 * i * vy0 - 1j * omega * es
)
dt_ed = (
vdc * i
- np.exp(-3 * 1j * theta) * np.conjugate(iss) * np.conjugate(vydelta)
- 2 * iss * vy0
- 2 * iss0 * vydelta
- 1j * omega * ed
)
dt_iss = 1 / Ind_sum * (vx - 1j * omega * Ind_sum * iss)
dt_iss0 = vx0 / Ind_sum
dt_i = 1 / L * (vy - (R + 1j * omega * L) * i - vg)
dt_theta = omega
dxx_dt_nv = [dt_es0, dt_ed0, dt_es, dt_ed, dt_iss, dt_iss0, dt_i, dt_theta]
return dxx_dt_nv
return rhs
import sys
import os
import numpy as np
import sympy as sp
import tabulate as tab
# tailing "_nv" stands for "numerical value"
model_name = "MMC"
# --------- CREATE SYMBOLIC PARAMETERS
pp_symb = [vdc, vg, omega, Lz, Mz, R, L] = sp.symbols("v_DC, v_g, omega, L_z, M_z, R, L", real=True)
# -------- CREATE AUXILIARY SYMBOLIC PARAMETERS
# (parameters, which shall not numerical represented in the parameter tabular)
# --------- SYMBOLIC PARAMETER FUNCTIONS
# ------------ parameter values can be constant/fixed values OR
# ------------ set in relation to other parameters (for example: a = 2*b)
# ------------ useful for a clean looking parameter table in the Documentation
# Due to performance of the simulation the parameters Lz, Mz and L are choosen to be scaled with 1/10
vdc_sf = 300
vg_sf = 235
omega_sf = 2 * sp.pi * 5
Lz_sf = 1.5 / 10
Mz_sf = 0.94 / 10
R_sf = 26
L_sf = 3 / 10
# List of symbolic parameter functions
pp_sf = [vdc_sf, vg_sf, omega_sf, Lz_sf, Mz_sf, R_sf, L_sf]
# Set numerical values of auxiliary parameters
# List for Substitution
# -- Entries are tuples like: (independent symbolic parameter, numerical value)
pp_subs_list = []
# OPTONAL: Dictionary which defines how certain variables shall be written
# in the tabular - key: Symbolic Variable, Value: LaTeX Representation/Code
# useful for example for complex variables: {Z: r"\underline{Z}"}
latex_names = {}
# ---------- CREATE BEGIN OF LATEX TABULAR
# Define tabular Header
# DON'T CHANGE FOLLOWING ENTRIES: "Symbol", "Value"
tabular_header = ["Parameter Name", "Symbol", "Value", "Unit"]
# Define column text alignments
col_alignment = ["left", "center", "left", "center"]
# Define Entries of all columns before the Symbol-Column
# --- Entries need to be latex code
col_1 = [
"DC voltage",
"grid voltage",
"angular speed",
"arm inductance",
"mutual inductance",
"load resistance",
"load inductance",
]
# contains all lists of the columns before the "Symbol" Column
# --- Empty list, if there are no columns before the "Symbol" Column
start_columns_list = [col_1]
# Define Entries of the columns after the Value-Column
# --- Entries need to be latex code
col_4 = ["V", "V", "Hz", "mH", "mH", r"$\Omega$", "mH"]
# contains all lists of columns after the FIX ENTRIES
# --- Empty list, if there are no columns after the "Value" column
end_columns_list = [col_4]
Related Problems:
Extensive Material:
Download pdf
Result: Success.
Last Build: Checkout CI Build
Runtime: 39.6 (estimated: 50s)
Plot:

The image of the latest CI job is not available. This is a fallback image.