Automatic Control Knowledge Repository
You currently have javascript disabled. Some features will be unavailable. Please consider enabling javascript.Details for: "Heat Equation"
Name: Heat Equation
(Key: 1VY9F)
Path: ackrep_data/system_models/heat_equation View on GitHub
Type: system_model
Short Description: PDE describing thermal conduction in matter
Created: 03.08.2022
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
Related Problems:
Extensive Material:
Download pdf
Result: Script Error.
Last Build: Checkout CI Build
Runtime: 2.4 (estimated: 20s)
The image of the latest CI job is not available. This is a fallback image.
The image of the latest CI job is not available. This is a fallback image. Debug:
Path: ackrep_data/system_models/heat_equation View on GitHub
Type: system_model
Short Description: PDE describing thermal conduction in matter
Created: 03.08.2022
Compatible Environment: default_conda_environment (Key: CDAMA)
Source Code [ / ] simulation.py
import numpy as np
import pyinduct as pi
import pyqtgraph as pg
import system_model
from scipy.integrate import solve_ivp
from ackrep_core import ResultContainer
from ackrep_core.system_model_management import save_plot_in_dir
import matplotlib.pyplot as plt
import matplotlib
import os
# link to documentation with examples: https://ackrep-doc.readthedocs.io/en/latest/devdoc/contributing_data.html
def simulate():
"""
simulate the system model with scipy.integrate.solve_ivp
:return: result of solve_ivp, might contains input function
"""
model = system_model.Model()
# simulation
def x0(z):
return 0 + model.y0 * z
start_func = pi.Function(x0, domain=model.spat_domain.bounds)
full_start_state = np.array([pi.project_on_base(start_func, pi.get_base("vis_base"))]).flatten()
initial_state = full_start_state[1:-1]
start_state_bar = model.a_tilde @ initial_state - (model.b1 * model.u(time=0)).flatten()
ss = pi.StateSpace(model.a_bar, model.b_bar, base_lbl="sim", input_handle=model.u)
sim_temp_domain, sim_weights_bar = pi.simulate_state_space(ss, start_state_bar, model.temp_domain)
# back-transformation
u_vec = np.reshape(model.u.get_results(sim_temp_domain), (len(model.temp_domain), 1))
sim_weights = sim_weights_bar @ model.a_tilde_inv + u_vec @ model.b1.T
# visualisation
plots = list()
save_pics = False
vis_weights = np.hstack((np.zeros_like(u_vec), sim_weights, u_vec))
eval_d = pi.evaluate_approximation("vis_base", vis_weights, sim_temp_domain, model.spat_domain, spat_order=0)
der_eval_d = pi.evaluate_approximation("vis_base", vis_weights, sim_temp_domain, model.spat_domain, spat_order=1)
pi.tear_down(("act_base", "sim_base", "vis_base"))
sim = ResultContainer()
sim.eval_d = eval_d
sim.der_eval_d = der_eval_d
sim.u = model.u
save_plot(sim)
return sim
def save_plot(simulation_data):
"""
plot your data and save the plot
access to data via: simulation_data.t array of time values
simulation_data.y array of data components
simulation_data.uu array of input values
:param simulation_data: simulation_data of system_model
:return: None
"""
# Note: plotting in pyinduct is usually done with pyqtgraph which causes issues during CI.
# This is why the plotting part doesnt look as clean.
# Pyinduct has its own plotting methods, feel free to use them in your own implementation.
matplotlib.use("Agg")
# input vis
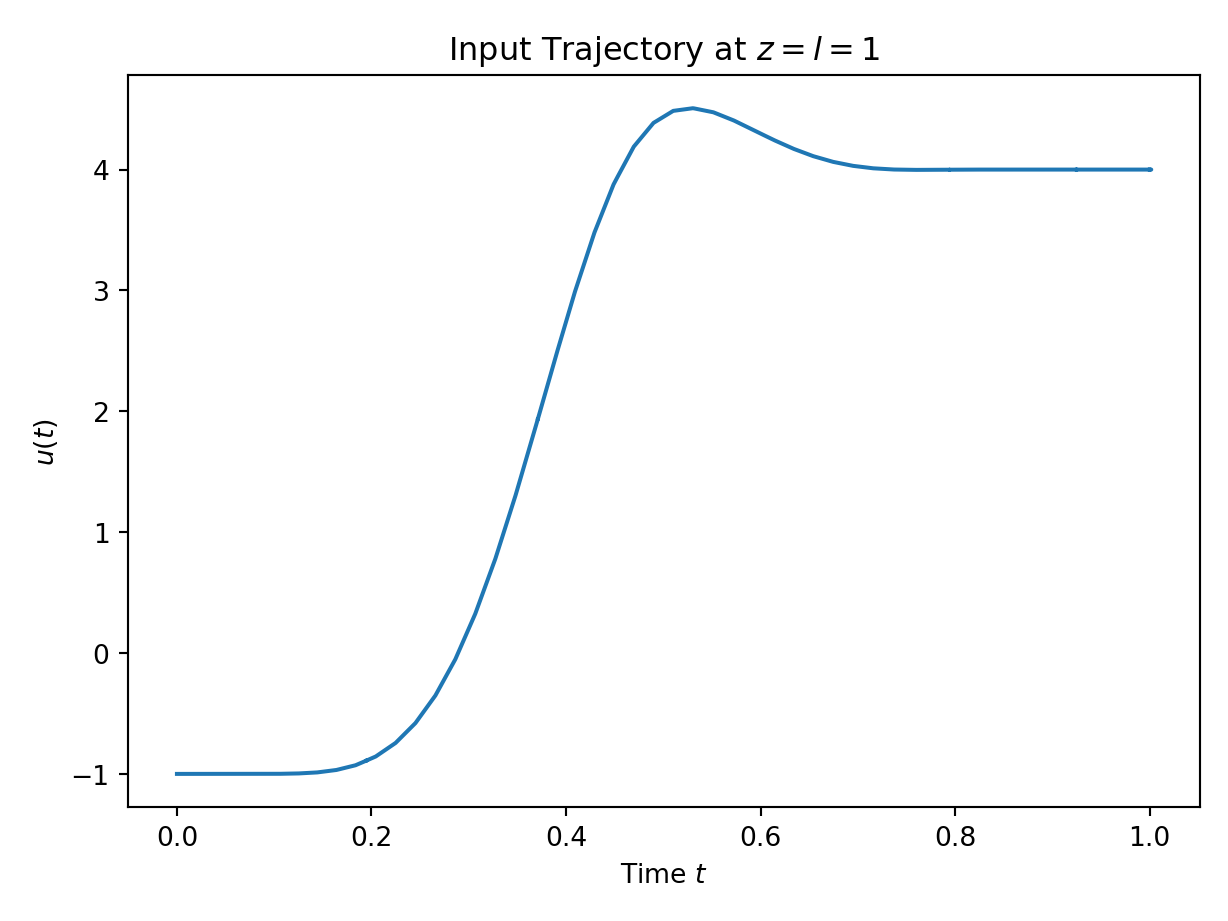
plt.plot(simulation_data.u._time_storage, simulation_data.u.get_results(simulation_data.u._time_storage))
plt.xlabel("Time $t$")
plt.ylabel("$u(t)$")
plt.title("Input Trajectory at $z=l=1$")
plt.grid()
plt.tight_layout()
save_plot_in_dir("plot_1.png")
win0 = pi.surface_plot(simulation_data.eval_d, zlabel="x(z,t)")
plt.title("Temperature Development in Time and Space")
plt.ylabel("Time $t$")
plt.xlabel("Space $z$")
plt.tight_layout()
save_plot_in_dir("plot_2.png")
# win2 = pi.PgAnimatedPlot(simulation_data.eval_d,
# labels=dict(left='x(z,t)', bottom='z'))
# win3 = pi.PgAnimatedPlot(simulation_data.der_eval_d,
# labels=dict(left='x\'(z,t)', bottom='z'))
# pi.show()
def evaluate_simulation(simulation_data):
"""
assert that the simulation results are as expected
:param simulation_data: simulation_data of system_model
:return:
"""
expected_final_state = np.array(
[
0.0,
0.02150636,
0.04301272,
0.06451907,
0.08602543,
0.10753179,
0.12903815,
0.1505445,
0.17205086,
0.19355722,
0.21506358,
0.23656994,
0.25807628,
0.27958259,
0.3010889,
0.32259522,
0.34410153,
0.36560784,
0.38711415,
0.40862047,
0.43012678,
0.45163309,
0.47313941,
0.49464572,
0.51615199,
0.53765824,
0.55916449,
0.58067075,
0.602177,
0.62368325,
0.64518951,
0.66669576,
0.68820201,
0.70970826,
0.73121452,
0.75272075,
0.77422688,
0.79573301,
0.81723913,
0.83874526,
0.86025138,
0.88175751,
0.90326363,
0.92476976,
0.94627588,
0.96778201,
0.98928813,
1.0107942,
1.03230022,
1.05380623,
1.07531225,
1.09681826,
1.11832428,
1.13983029,
1.16133631,
1.18284232,
1.20434834,
1.22585435,
1.24736037,
1.26886621,
1.29037204,
1.31187786,
1.33338368,
1.35488951,
1.37639533,
1.39790116,
1.41940698,
1.4409128,
1.46241863,
1.48392445,
1.50543024,
1.52693592,
1.5484416,
1.56994728,
1.59145296,
1.61295863,
1.63446431,
1.65596999,
1.67747567,
1.69898135,
1.72048703,
1.74199271,
1.76349825,
1.7850037,
1.80650916,
1.82801461,
1.84952007,
1.87102552,
1.89253097,
1.91403643,
1.93554188,
1.95704734,
1.97855279,
2.00005824,
2.02156354,
2.04306884,
2.06457414,
2.08607943,
2.10758473,
2.12909003,
2.15059533,
2.17210063,
2.19360592,
2.21511122,
2.23661652,
2.25812173,
2.27962681,
2.30113188,
2.32263695,
2.34414202,
2.3656471,
2.38715217,
2.40865724,
2.43016232,
2.45166739,
2.47317246,
2.49467753,
2.5161825,
2.53768743,
2.55919236,
2.58069728,
2.60220221,
2.62370714,
2.64521207,
2.666717,
2.68822193,
2.70972686,
2.73123178,
2.75273669,
2.77424143,
2.79574616,
2.8172509,
2.83875564,
2.86026038,
2.88176512,
2.90326985,
2.92477459,
2.94627933,
2.96778407,
2.9892888,
3.01079349,
3.03229811,
3.05380274,
3.07530737,
3.096812,
3.11831662,
3.13982125,
3.16132588,
3.18283051,
3.20433513,
3.22583976,
3.24734439,
3.26884891,
3.29035341,
3.31185791,
3.33336241,
3.35486691,
3.37637141,
3.39787591,
3.41938041,
3.44088491,
3.46238941,
3.48389391,
3.50539839,
3.52690283,
3.54840727,
3.56991171,
3.59141615,
3.61292059,
3.63442503,
3.65592947,
3.67743391,
3.69893835,
3.72044279,
3.74194724,
3.76345165,
3.78495604,
3.80646044,
3.82796483,
3.84946923,
3.87097363,
3.89247802,
3.91398242,
3.93548681,
3.95699121,
3.9784956,
4.0,
]
)
rc = ResultContainer(score=1.0)
simulated_final_state = simulation_data.eval_d.output_data[-1]
rc.final_state_errors = [
simulated_final_state[i] - expected_final_state[i] for i in np.arange(0, len(simulated_final_state))
]
rc.success = np.allclose(expected_final_state, simulated_final_state, rtol=0, atol=1e-2)
return rc
import numpy as np
import pyinduct as pi
import pyinduct.parabolic as parabolic
import matplotlib.pyplot as plt
from ipydex import IPS
def run(show_plots):
n_fem = 17
T = 1
l = 1
y0 = -1
y1 = 4
param = [1, 0, 0, None, None]
# or try these:
# param = [1, -0.5, -8, None, None] # :)))
a2, a1, a0, _, _ = param
temp_domain = pi.Domain(bounds=(0, T), num=100)
spat_domain = pi.Domain(bounds=(0, l), num=n_fem * 11)
# initial and test functions
nodes = pi.Domain(spat_domain.bounds, num=n_fem)
fem_base = pi.LagrangeFirstOrder.cure_interval(nodes)
act_fem_base = pi.Base(fem_base[-1])
not_act_fem_base = pi.Base(fem_base[1:-1])
vis_fems_base = pi.Base(fem_base)
pi.register_base("act_base", act_fem_base)
pi.register_base("sim_base", not_act_fem_base)
pi.register_base("vis_base", vis_fems_base)
# trajectory
u = parabolic.RadFeedForward(
l, T, param_original=param, bound_cond_type="dirichlet", actuation_type="dirichlet", y_start=y0, y_end=y1
)
# weak form
x = pi.FieldVariable("sim_base")
x_dt = x.derive(temp_order=1)
x_dz = x.derive(spat_order=1)
phi = pi.TestFunction("sim_base")
phi_dz = phi.derive(1)
act_phi = pi.ScalarFunction("act_base")
act_phi_dz = act_phi.derive(1)
# weak formulation of the PDE equations (see documentation)
weak_form = pi.WeakFormulation(
[
# ... of the homogeneous part of the system
pi.IntegralTerm(pi.Product(x_dt, phi), limits=spat_domain.bounds),
pi.IntegralTerm(pi.Product(x_dz, phi_dz), limits=spat_domain.bounds, scale=a2),
pi.IntegralTerm(pi.Product(x_dz, phi), limits=spat_domain.bounds, scale=-a1),
pi.IntegralTerm(pi.Product(x, phi), limits=spat_domain.bounds, scale=-a0),
# ... of the inhomogeneous part of the system
pi.IntegralTerm(pi.Product(pi.Product(act_phi, phi), pi.Input(u, order=1)), limits=spat_domain.bounds),
pi.IntegralTerm(
pi.Product(pi.Product(act_phi_dz, phi_dz), pi.Input(u)), limits=spat_domain.bounds, scale=a2
),
pi.IntegralTerm(pi.Product(pi.Product(act_phi_dz, phi), pi.Input(u)), limits=spat_domain.bounds, scale=-a1),
pi.IntegralTerm(pi.Product(pi.Product(act_phi, phi), pi.Input(u)), limits=spat_domain.bounds, scale=-a0),
],
name="main_system",
)
# system matrices \dot x = A x + b0 u + b1 \dot u
cf = pi.parse_weak_formulation(weak_form)
ss = pi.create_state_space(cf)
a_mat = ss.A[1]
b0 = ss.B[0][1]
b1 = ss.B[1][1]
# transformation into \dot \bar x = \bar A \bar x + \bar b u
a_tilde = np.diag(np.ones(a_mat.shape[0]), 0)
a_tilde_inv = np.linalg.inv(a_tilde)
a_bar = (a_tilde @ a_mat) @ a_tilde_inv
b_bar = a_tilde @ (a_mat @ b1) + b0
# simulation
def x0(z):
return 0 + y0 * z
start_func = pi.Function(x0, domain=spat_domain.bounds)
full_start_state = np.array([pi.project_on_base(start_func, pi.get_base("vis_base"))]).flatten()
initial_state = full_start_state[1:-1]
start_state_bar = a_tilde @ initial_state - (b1 * u(time=0)).flatten()
ss = pi.StateSpace(a_bar, b_bar, base_lbl="sim", input_handle=u)
sim_temp_domain, sim_weights_bar = pi.simulate_state_space(ss, start_state_bar, temp_domain)
# back-transformation
u_vec = np.reshape(u.get_results(sim_temp_domain), (len(temp_domain), 1))
sim_weights = sim_weights_bar @ a_tilde_inv + u_vec @ b1.T
# visualisation
plots = list()
save_pics = False
vis_weights = np.hstack((np.zeros_like(u_vec), sim_weights, u_vec))
eval_d = pi.evaluate_approximation("vis_base", vis_weights, sim_temp_domain, spat_domain, spat_order=0)
der_eval_d = pi.evaluate_approximation("vis_base", vis_weights, sim_temp_domain, spat_domain, spat_order=1)
if show_plots:
plots.append(pi.PgAnimatedPlot(eval_d,
labels=dict(left='x(z,t)', bottom='z'),
save_pics=save_pics))
# plots.append(pi.PgAnimatedPlot(der_eval_d,
# labels=dict(left='x\'(z,t)', bottom='z'),
# save_pics=save_pics))
win1 = pi.surface_plot(eval_d, zlabel="x(z,t)")
# win2 = pi.surface_plot(der_eval_d, title="x'(z,t)")
# win1.gl_widget.grabFrameBuffer().save(path + "rad_dirichlet_3d_x.png")
# win2.gl_widget.grabFrameBuffer().save(path + 'rad_dirichlet_3d_dx.png')
# pi.show()
plt.show()
pi.tear_down(("act_base", "sim_base", "vis_base"))
if __name__ == "__main__":
run(True)
import sympy as sp
import symbtools as st
import importlib
import sys, os
import pyinduct as pi
import pyinduct.parabolic as parabolic
import numpy as np
from ipydex import IPS, activate_ips_on_exception
from ackrep_core.system_model_management import GenericModel, import_parameters
class Model:
# Import parameter_file
params = import_parameters()
a2 = [float(i[1]) for i in params.get_default_parameters().items()][0]
n_fem = 17
l = 1
T = 1
# start and end of input trajectory
y0 = -1
y1 = 4
# coefs of pde
coefs = [1, 0, 0, None, None]
# or try these:
# coefs = [1, -0.5, -8, None, None]
_, a1, a0, _, _ = coefs
temp_domain = pi.Domain(bounds=(0, T), num=100)
spat_domain = pi.Domain(bounds=(0, l), num=n_fem * 11)
# initial and test functions
nodes = pi.Domain(spat_domain.bounds, num=n_fem)
fem_base = pi.LagrangeFirstOrder.cure_interval(nodes)
act_fem_base = pi.Base(fem_base[-1])
not_act_fem_base = pi.Base(fem_base[1:-1])
vis_fems_base = pi.Base(fem_base)
pi.register_base("act_base", act_fem_base)
pi.register_base("sim_base", not_act_fem_base)
pi.register_base("vis_base", vis_fems_base)
# trajectory
u = parabolic.RadFeedForward(
l, T, param_original=coefs, bound_cond_type="dirichlet", actuation_type="dirichlet", y_start=y0, y_end=y1
)
# weak form
x = pi.FieldVariable("sim_base")
x_dt = x.derive(temp_order=1)
x_dz = x.derive(spat_order=1)
phi = pi.TestFunction("sim_base")
phi_dz = phi.derive(1)
act_phi = pi.ScalarFunction("act_base")
act_phi_dz = act_phi.derive(1)
# weak formulation of the PDE equations (see documentation)
weak_form = pi.WeakFormulation(
[
# ... of the homogeneous part of the system
pi.IntegralTerm(pi.Product(x_dt, phi), limits=spat_domain.bounds),
pi.IntegralTerm(pi.Product(x_dz, phi_dz), limits=spat_domain.bounds, scale=a2),
pi.IntegralTerm(pi.Product(x_dz, phi), limits=spat_domain.bounds, scale=-a1),
pi.IntegralTerm(pi.Product(x, phi), limits=spat_domain.bounds, scale=-a0),
# ... of the inhomogeneous part of the system
pi.IntegralTerm(pi.Product(pi.Product(act_phi, phi), pi.Input(u, order=1)), limits=spat_domain.bounds),
pi.IntegralTerm(
pi.Product(pi.Product(act_phi_dz, phi_dz), pi.Input(u)), limits=spat_domain.bounds, scale=a2
),
pi.IntegralTerm(pi.Product(pi.Product(act_phi_dz, phi), pi.Input(u)), limits=spat_domain.bounds, scale=-a1),
pi.IntegralTerm(pi.Product(pi.Product(act_phi, phi), pi.Input(u)), limits=spat_domain.bounds, scale=-a0),
],
name="main_system",
)
# system matrices \dot x = A x + b0 u + b1 \dot u
cf = pi.parse_weak_formulation(weak_form)
ss = pi.create_state_space(cf)
a_mat = ss.A[1]
b0 = ss.B[0][1]
b1 = ss.B[1][1]
# transformation into \dot \bar x = \bar A \bar x + \bar b u
a_tilde = np.diag(np.ones(a_mat.shape[0]), 0)
a_tilde_inv = np.linalg.inv(a_tilde)
a_bar = (a_tilde @ a_mat) @ a_tilde_inv
b_bar = a_tilde @ (a_mat @ b1) + b0
# -*- coding: utf-8 -*-
"""
Created on Fri Jun 11 13:51:06 2021
@author: Jonathan Rockstroh
"""
import sys
import os
import numpy as np
import sympy as sp
import tabulate as tab
# tailing "_nv" stands for "numerical value"
model_name = "Heat_Equation"
# CREATE SYMBOLIC PARAMETERS
pp_symb = [alpha] = [sp.symbols("alpha", real=True)]
# SYMBOLIC PARAMETER FUNCTIONS
alpha_sf = 1
# List of symbolic parameter functions
pp_sf = [alpha_sf]
# List for Substitution
pp_subs_list = []
# OPTONAL: Dictionary which defines how certain variables shall be written
# in the tabular - key: Symbolic Variable, Value: LaTeX Representation/Code
# useful for example for complex variables: {Z: r"\underline{Z}"}
latex_names = {alpha: r"\alpha"}
# ---------- CREATE BEGIN OF LATEX TABULAR
# Define tabular Header
# DON'T CHANGE FOLLOWING ENTRIES: "Symbol", "Value"
tabular_header = ["Parameter Name", "Symbol", "Value"]
# Define column text alignments
col_alignment = ["left", "center", "center"]
# Define Entries of all columns before the Symbol-Column
# --- Entries need to be latex code
col_1 = ["thermal diffusivity"]
# contains all lists of the columns before the "Symbol" Column
# --- Empty list, if there are no columns before the "Symbol" Column
start_columns_list = [col_1]
# contains all lists of columns after the FIX ENTRIES
# --- Empty list, if there are no columns after the "Value" column
end_columns_list = []
Related Problems:
Extensive Material:
Download pdf
Result: Script Error.
Last Build: Checkout CI Build
Runtime: 2.4 (estimated: 20s)
Entity passed last: 2023-07-03 13:14:44
with ackrep_data commit: {'author': 'JuliusFiedler', 'branch': 'develop', 'date': '2023-07-03 13:12:35', 'message': 'fix typos and folder names', 'sha': '180d385f485e4aa0d3df6c7a716fa9d1270dc4f3'}
with ackrep_core commit: {'author': 'JuliusFiedler', 'branch': 'develop', 'date': '2023-07-03 09:44:41', 'message': 'black', 'sha': '2bde08d26afc7227546533ecd58f249d24063462'}
in environment: default_conda_environment:0.1.12
Checkout CI Build
Plot:with ackrep_data commit: {'author': 'JuliusFiedler', 'branch': 'develop', 'date': '2023-07-03 13:12:35', 'message': 'fix typos and folder names', 'sha': '180d385f485e4aa0d3df6c7a716fa9d1270dc4f3'}
with ackrep_core commit: {'author': 'JuliusFiedler', 'branch': 'develop', 'date': '2023-07-03 09:44:41', 'message': 'black', 'sha': '2bde08d26afc7227546533ecd58f249d24063462'}
in environment: default_conda_environment:0.1.12
Checkout CI Build

The image of the latest CI job is not available. This is a fallback image.

The image of the latest CI job is not available. This is a fallback image. Debug:
Checking <SystemModel (pk: 134, key: 1VY9F)> "(Heat Equation, 20s)"
13:02:25 - ackrep - INFO - ... Creating exec-script ...
13:02:25 - ackrep - INFO - execscript-path: /code/ackrep/ackrep_data/execscript.py
13:02:25 - ackrep - INFO - ... running exec-script /code/ackrep/ackrep_data/execscript.py ...
13:02:27 - ackrep - ERROR -
The command `python /code/ackrep/ackrep_data/execscript.py` exited with returncode 1.
stdout:
stderr: Traceback (most recent call last):
File "/code/ackrep/ackrep_data/execscript.py", line 35, in <module>
import system_model
File "/code/ackrep/ackrep_data/system_models/heat_equation/system_model.py", line 5, in <module>
import pyinduct as pi
File "/opt/conda/lib/python3.8/site-packages/pyinduct/__init__.py", line 17, in <module>
from .eigenfunctions import *
File "/opt/conda/lib/python3.8/site-packages/pyinduct/eigenfunctions.py", line 24, in <module>
from .visualization import visualize_roots
File "/opt/conda/lib/python3.8/site-packages/pyinduct/visualization.py", line 19, in <module>
import pyqtgraph.opengl as gl
File "/opt/conda/lib/python3.8/site-packages/pyqtgraph/opengl/__init__.py", line 1, in <module>
from . import shaders
File "/opt/conda/lib/python3.8/site-packages/pyqtgraph/opengl/shaders.py", line 1, in <module>
from OpenGL.GL import * # noqa
File "/opt/conda/lib/python3.8/site-packages/OpenGL/GL/__init__.py", line 4, in <module>
from OpenGL.GL.VERSION.GL_1_1 import *
File "/opt/conda/lib/python3.8/site-packages/OpenGL/GL/VERSION/GL_1_1.py", line 14, in <module>
from OpenGL.raw.GL.VERSION.GL_1_1 import *
File "/opt/conda/lib/python3.8/site-packages/OpenGL/raw/GL/VERSION/GL_1_1.py", line 7, in <module>
from OpenGL.raw.GL import _errors
File "/opt/conda/lib/python3.8/site-packages/OpenGL/raw/GL/_errors.py", line 4, in <module>
_error_checker = _ErrorChecker( _p, _p.GL.glGetError )
AttributeError: 'NoneType' object has no attribute 'glGetError'
Fail.